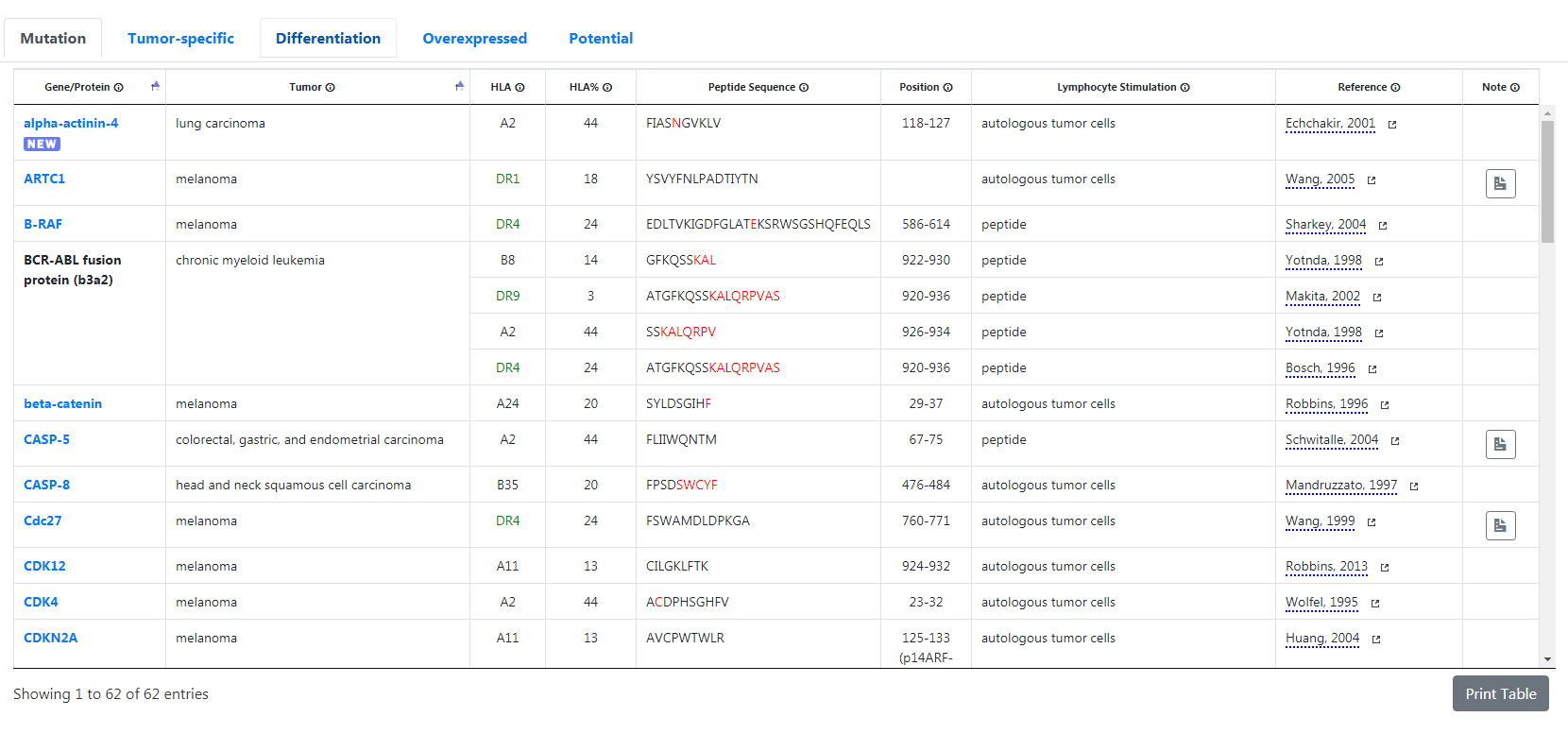
Database page
The Database page displays tables of peptides recorded in the database. Click on the desired tab to display the table of your choice.
- The “mutation” tab will display unique antigens which result from point mutations in genes that are expressed ubiquitously.
- The “tumor-specific” tab will display shared antigens which are expressed in many tumors but not in normal tissues.
- The “differentiation” tab will display shared antigens which are also expressed in the normal tissue of origin of the malignancy.
- The “overexpressed” tab will display shared antigens which are expressed in a wide variety of normal tissues and overexpressed in tumors.
- Finally, the “potential” tab will display references of articles that have not (yet) been included in the tables because formal evidence to fulfill one or several of the aforementioned criteria has not been provided.
Metadata
Every table except the "potential" table has several columns with the following metadata:
- Gene/Protein: The name of the gene or protein. If a geneCard url is available, you can access it by clicking on it. Newly added genes will have a
 icon.
icon. - Normal/Tumor: Depending of the gene table, this column indicates the type of tissue or tumor which expresses the (mutated) gene. You can click on the
 icon to display the expression profile in the tissues.
icon to display the expression profile in the tissues. - HLA: Human leukocyte antigen. Please note class II HLA are shown in green.
- HLA%: Frequency in Caucasians based on based on (1) Marsh SGE, Parham P, Barber LD. The HLA Factsbook (Academic Press, 2000) for HLA-A, B, C, and (2) Colombani J. HLA, Fonctions immunitaires et applications médicales (John Libbey Eurotext, 1993) for HLA-DP and DR.
- Peptide sequence. Modified residues resulting from mutations in unique antigens are indicated in red.
- Position: Position of the peptide sequence in the protein sequence
- Lymphocyte stimulation: Method used to isolate the CTL recognizing the antigen
- Reference: The first author and the year of publication. If you click on dotted text, the complete reference will be displayed at the bottom of the table (it will be cleared if you change the table tab or if you scroll up or down the table). If you click on the
 icon, your browser will open a new window or tab with the PubMed url
icon, your browser will open a new window or tab with the PubMed url - Note: Comment indicating specific characteristics of the gene, e.g. result of frameshift mutation etc. Hover over the icon to display the comment.

Search page
The search page is designed to help you browse through the complete database with ease.
Use the navigation buttons on the top left to find your desired type of gene immediately. Results can be sorted by clicking on the column headers and multiple columns can be sorted by holding the shift key before clicking on the columns you want to sort together. In such case, the order of the columns you clicked will determine the order of the sort process.
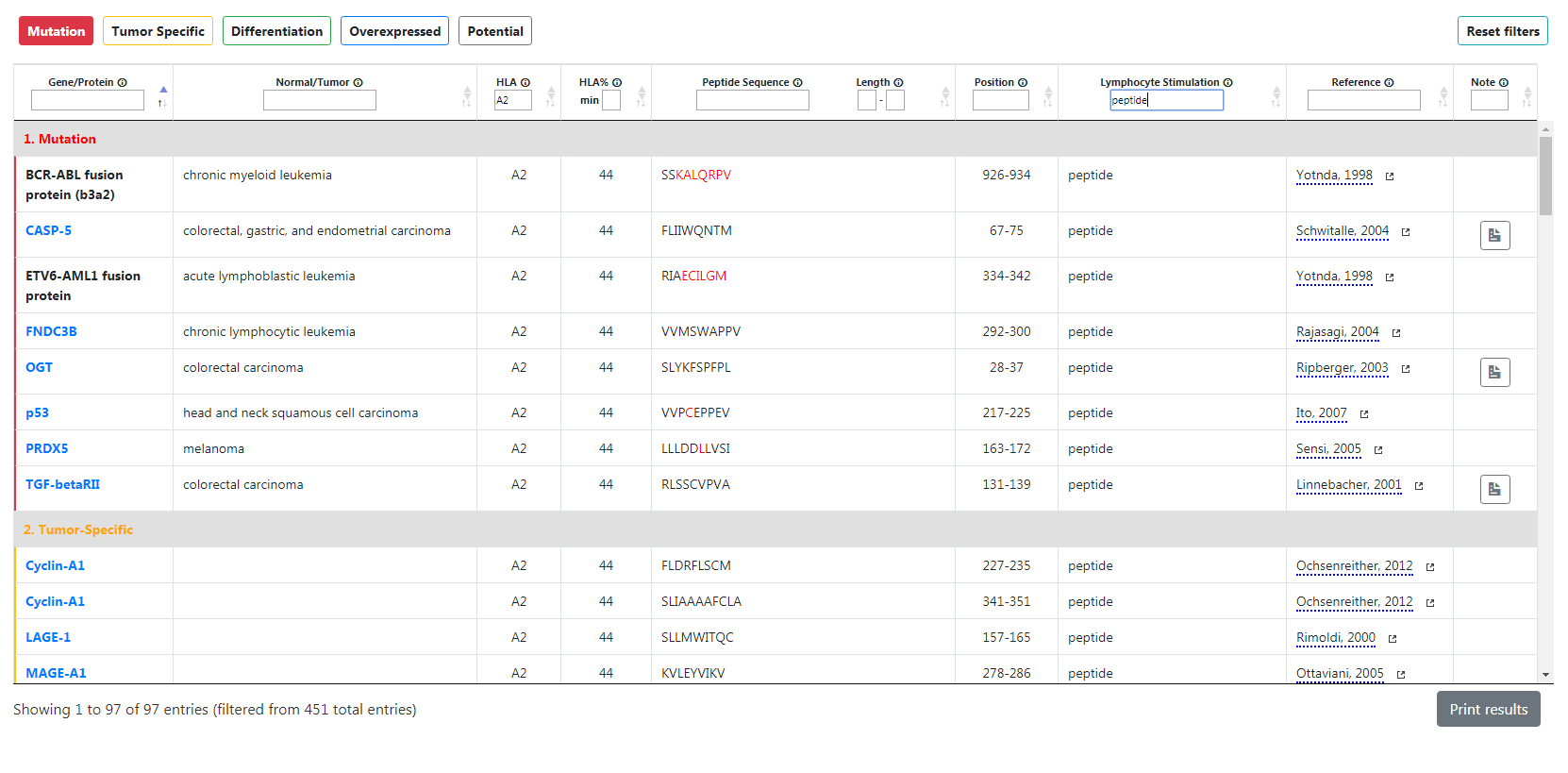
Of course, you can filter the table so you can search the genes for specific criteria. All you have to do is to use the filters located below the column headers. Any input in those fields will immediately filter the results from the search table without any need of reloading the page. You can also combine several filters for a more precise search.
For instance, if you type “A2” in the HLA field and “peptide” in the lymphocyte stimulation field, only genes having an HLA and lymphocyte stimulation matching those criteria will be displayed. In that example, there are only 97 entries that match both critera. The number of entries based on your search criteria is displayed at the bottom left of the table.
Reset your search criteria and the whole table with the Reset Filters button on the top right.
Please note that any filter aside of reference will clear the potential table as its content is not relevant to the metadata you are looking for.
General filters
With the exception of the HLA field, searches done using the filter fields will display genes having metadata containing the letters/words you have typed (lower and upper case won't matter). You do not have to type the exact terms.
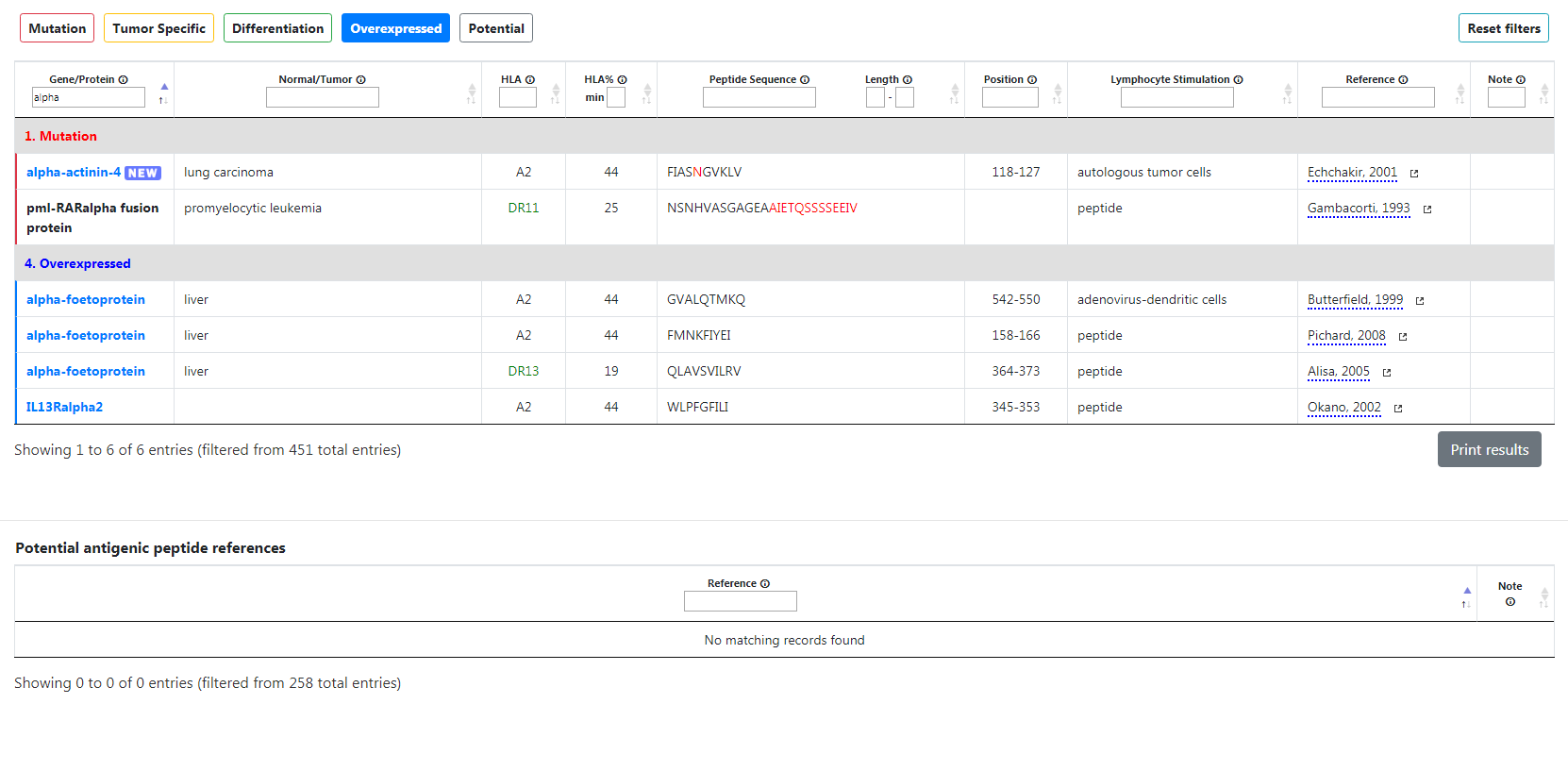
For example, if you type “alpha” in the Gene/Protein filter, it will show every gene with “alpha” in their name, such as alpha-actinin-4 or pml-RARalpha fusion protein. However, please note that you cannot combine several terms within the same filter. For example, if you type “alpha RAR”, it will not show pml-RARalpha fusion protein because the filter will not search for both “alpha” and “RAR” but rather one single word “alpha RAR”. Please note the HLA column is different and requires accurate search input. For instance, if you type "A2", the table will not show genes with an "A22" HLA.
Reference filter
The reference filter will apply to the complete reference of the article related to the gene. As such, you can type the name of any author, the year when the article was published or the title of the article.
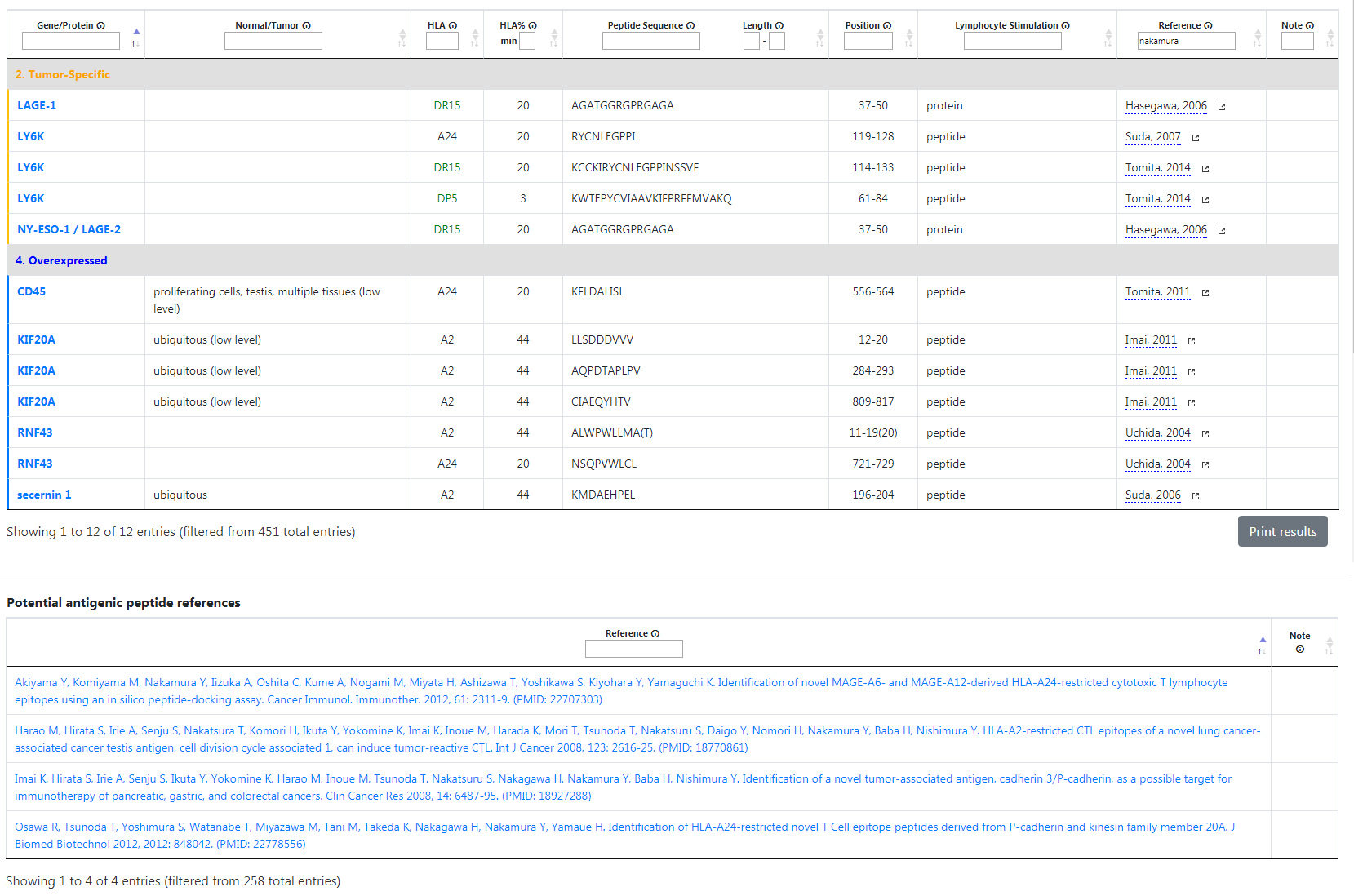
For example, if you type "Nakamura" in the reference filter, the table will display 12 results since Mr. Nakamura was a co-author of the articles of those genes.
Unlike other filters, the reference filter will also apply to the potential table as well. In this example, 4 entries are displayed, since Mr. Nakamura is one of the listed authors of those articles.
Wildcard
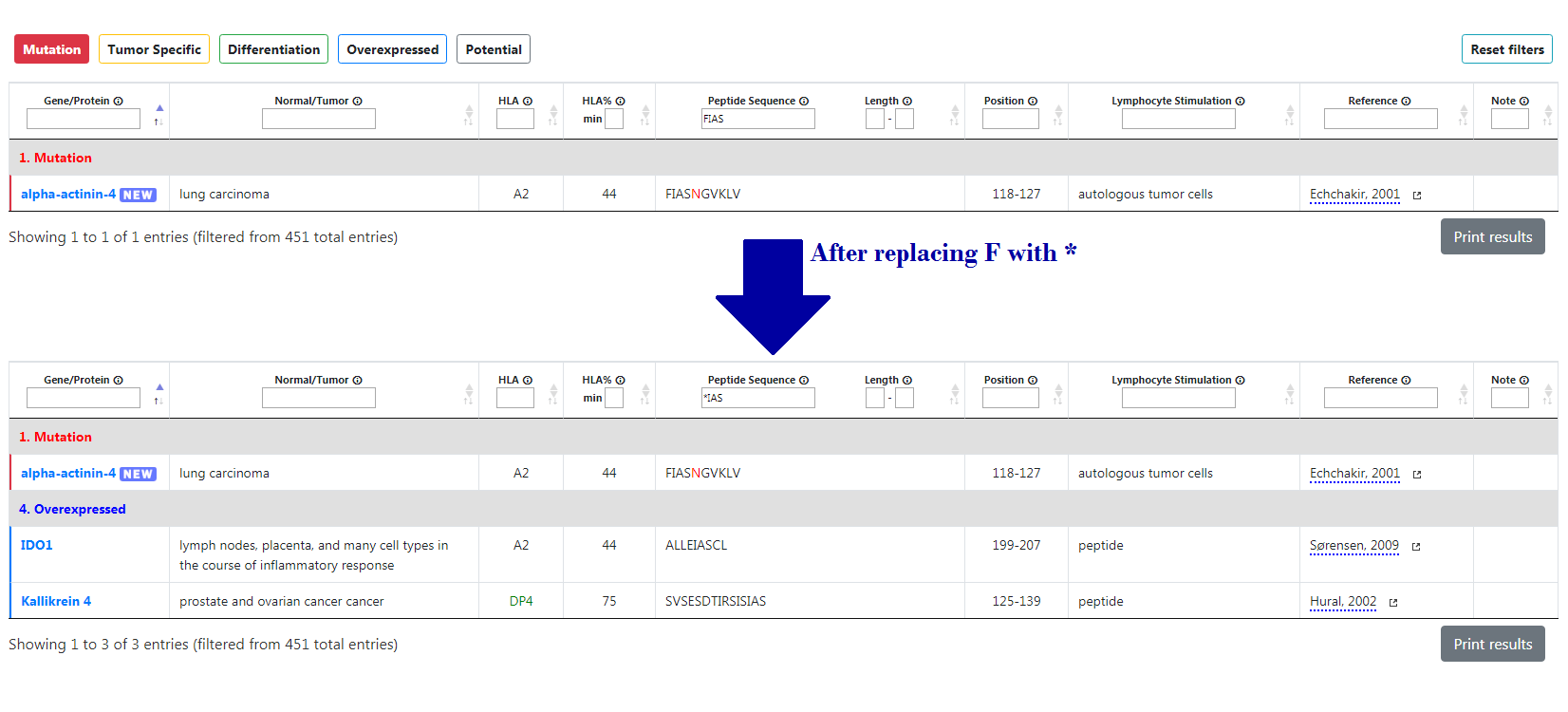
Use "*" as a wildcard, which replaces any 1 character or none. Several wildcards can be used at once, in which case the search function will consider any combination of letters equal or shorter than the number of * put together.
For instance, if you type “FIAS” in the peptide sequence filter, this will only display alpha-actinin-4. On the other hand, if you type “*IAS”, it will yield more results.
Using the previous HLA example, if you type "A2*", it will display any gene with an "A2" HLA, "A24" HLA etc.
In the same example with multiple wildcards, "A**" will lead to "A1", "A2", "A11" etc. In other words, ** is any combination of 0 to 2 letters together.
Delimiters
Furthermore, you can also use "(" and ")" to delimit your search: any filter input starting with "(" means that the genes must have the same first letters in the relevant column. Likewise, any filter input ending with ")" means that the last letters must match them.
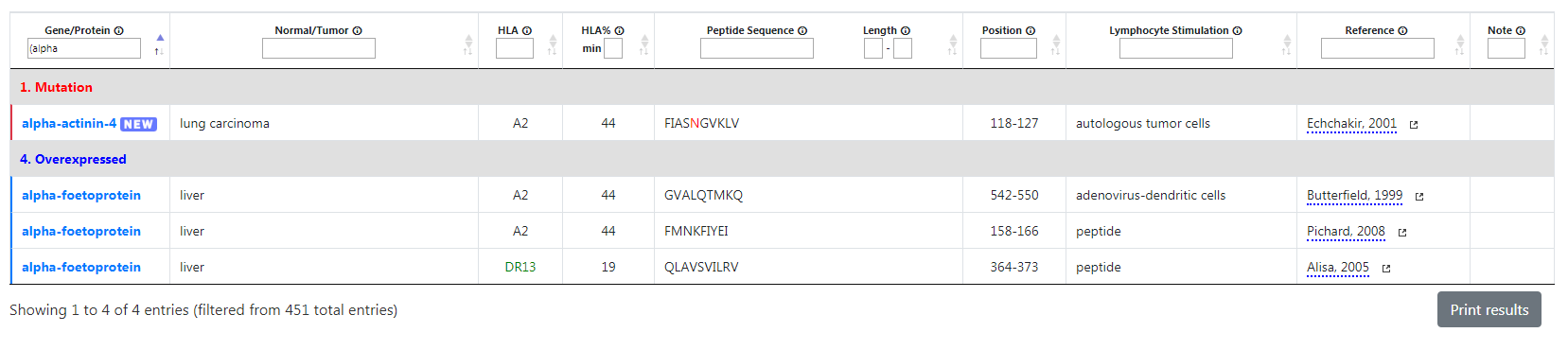
Example: if you type “(alpha” in the Gene/Protein filter only alpha-actinin-4 and alpha-foetoprotein will be displayed as the other genes names do not start with “alpha”.
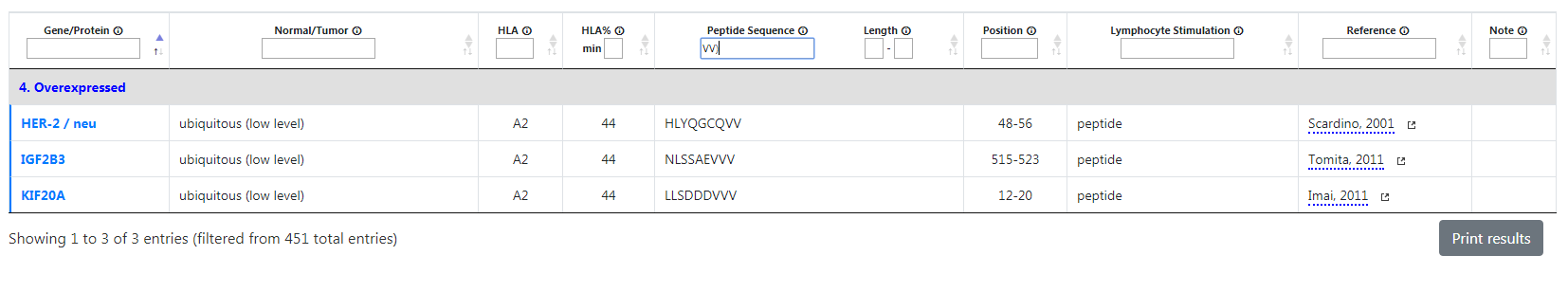
Similarly, if you type “VV)” in the peptide sequence filter, only HER-2, IGF2B3 and KIF20A will be shown. Other genes with VV won't be displayed if VV aren't the last letters of their sequence.
Example: Search page with Peptide sequence set to VV)